4. Full-cell validation¶
In this example, we load the previous results from OCP fitting, electrode balancing, and half-cell GITT fitting to create a parameter set for the full cell. We then validate this parameter set against the real parameters.
from pathlib import Path
import ionworkspipeline as iwp
import pybamm
from plots import constant_current, drive_cycle
from true_parameters.parameters import full_cell
We begin by making a list of “known parameters”. These are values that are already known from other sources such as spec sheets, direct measurement, and/or other experiments. For this example, we load the known parameters from the true parameters used to generate the synthetic data.
# Known values
KNOWN_PARAMETERS = [
# Negative electrode
"Negative electrode thickness [m]",
"Negative electrode conductivity [S.m-1]",
"Negative electrode porosity",
"Negative electrode active material volume fraction",
"Negative particle radius [m]",
"Negative electrode OCP entropic change [V.K-1]",
# Separator
"Separator thickness [m]",
"Separator porosity",
# Positive electrode
"Positive electrode thickness [m]",
"Positive electrode conductivity [S.m-1]",
"Positive electrode porosity",
"Positive electrode active material volume fraction",
"Positive particle radius [m]",
"Positive electrode OCP entropic change [V.K-1]",
# Cell
"Electrode height [m]",
"Electrode width [m]",
# these are user-specified at the cell level and may differ from the actual
# voltage and capacity as measured in the experiment
"Lower voltage cut-off [V]",
"Upper voltage cut-off [V]",
"Open-circuit voltage at 0% SOC [V]",
"Open-circuit voltage at 100% SOC [V]",
"Nominal cell capacity [A.h]",
"Current function [A]",
]
Next we load in the known parameters
true_parameter_values = full_cell()
known_parameter_values = {k: true_parameter_values[k] for k in KNOWN_PARAMETERS}
known_values = iwp.direct_entries.DirectEntry(
known_parameter_values, "Measured or assumed values"
)
and load in the previously fitted open circuit, electrode balancing and GITT parameters.
ocp_n = iwp.calculations.ocp_data_interpolant_from_csv(
"negative",
"fitted_parameters/negative_electrode_ocp.csv",
)
ocp_p = iwp.calculations.ocp_data_interpolant_from_csv(
"positive",
"fitted_parameters/positive_electrode_ocp.csv",
)
electrode_balance_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "full_cell_balance.json",
)
def j0_n(c_e, c_s_surf, c_s_max, T):
j0_ref = pybamm.Parameter(
"Negative electrode reference exchange-current density [A.m-2]"
)
alpha = 0.5
c_e_ref = 1000
return (
j0_ref
* (c_e / c_e_ref) ** (1 - alpha)
* (c_s_surf / c_s_max) ** alpha
* (1 - c_s_surf / c_s_max) ** (1 - alpha)
)
def j0_p(c_e, c_s_surf, c_s_max, T):
j0_ref = pybamm.Parameter(
"Positive electrode reference exchange-current density [A.m-2]"
)
alpha = 0.5
c_e_ref = 1000
return (
j0_ref
* (c_e / c_e_ref) ** (1 - alpha)
* (c_s_surf / c_s_max) ** alpha
* (1 - c_s_surf / c_s_max) ** (1 - alpha)
)
exchange_current_params = iwp.direct_entries.DirectEntry(
{
"Negative electrode exchange-current density [A.m-2]": j0_n,
"Positive electrode exchange-current density [A.m-2]": j0_p,
},
source="Known functional forms for exchange-current densities",
)
negative_gitt_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "negative_electrode_gitt.json",
)
positive_gitt_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "positive_electrode_gitt.json",
)
Next, we construct a pipeline that uses the known parameters and the fitted parameters to create a full cell parameter set.
pipeline = iwp.Pipeline(
{
"known values": known_values,
"negative OCP": ocp_n,
"positive OCP": ocp_p,
"Electrode balance parameters": electrode_balance_params,
"Exchange-current densities": exchange_current_params,
"Negative GITT parameters": negative_gitt_params,
"Positive GITT parameters": positive_gitt_params,
"Defaults": iwp.direct_entries.standard_defaults(),
"Bruggeman": iwp.direct_entries.bruggeman(),
"Temperatures": iwp.direct_entries.temperatures(298.15),
"Electrolyte": iwp.direct_entries.nyman_electrolyte(1000),
"Maximum concentration negative": iwp.calculations.ElectrodeCapacity(
"negative",
unknown="maximum concentration",
method="capacity",
),
"Maximum concentration positive": iwp.calculations.ElectrodeCapacity(
"positive",
unknown="maximum concentration",
method="capacity",
),
"Initial concentration negative": iwp.calculations.InitialSOCHalfCell(
"negative", "maximum"
),
"Initial concentration positive": iwp.calculations.InitialSOCHalfCell(
"positive", "maximum"
),
}
)
We run the pipeline
fitted_parameter_values = pipeline.run()
and export a python script for the full cell parameter values
# Export a .py file with the PyBaMM parameters
# Note: if we specify data_path=None, the variable DATA_PATH will be set to the
# directory containing the exported script
iwp.util.export_python_script(
fitted_parameter_values,
"fitted_parameters/full_cell.py",
data_path=None,
)
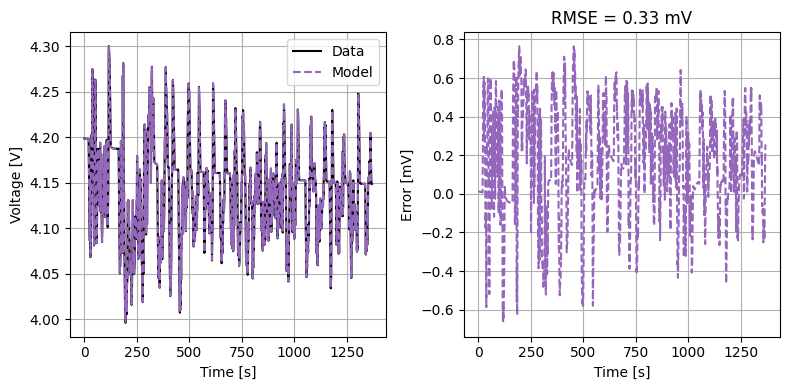
Finally, we validate this parameter set against the known parameters.
model = pybamm.lithium_ion.SPMe()
true_parameter_values = pybamm.ParameterValues(true_parameter_values)
def plot_constant_current(model, Crate=1):
constant_current(
model,
true_parameter_values,
fitted_parameter_values,
Crate=Crate,
initial_soc=1,
)
def plot_drive_cycle(model, peak_current=None):
peak_current = peak_current or true_parameter_values["Nominal cell capacity [A.h]"]
drive_cycle(
model,
true_parameter_values,
fitted_parameter_values,
peak_current=peak_current,
initial_soc=1,
)
plot_constant_current(model, Crate=1)
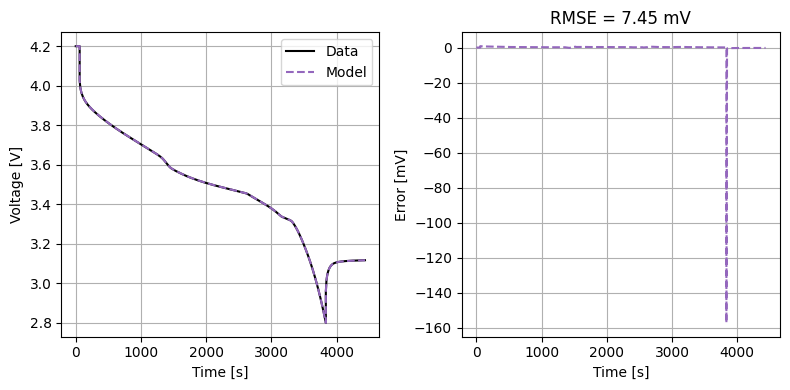
plot_drive_cycle(model)