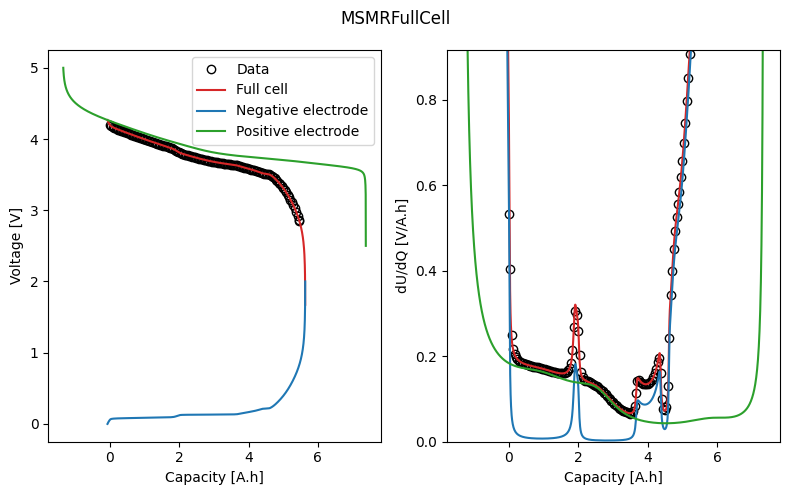
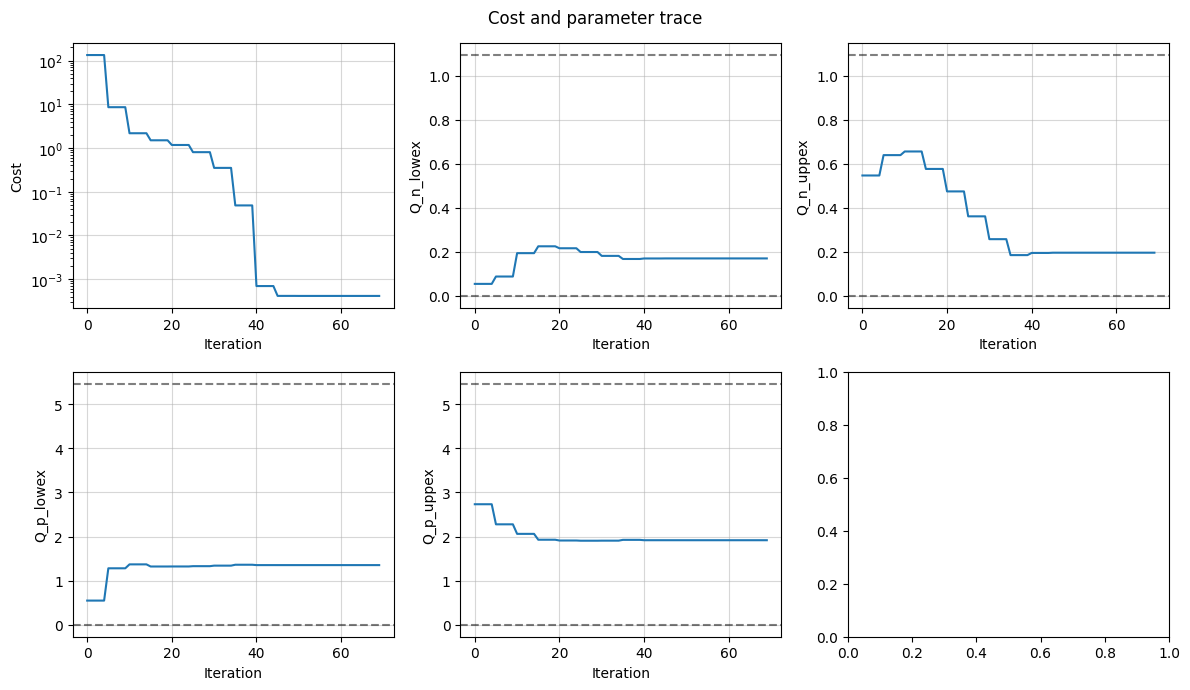
2. Full-cell OCV¶
In this example, we fit the electrode balance for a full-cell, using the half-cell MSMR functions fitted and saved in the previous notebook.
Before running this example, make sure to run the script true_parameters/generate_data.py to generate the synthetic data.
from pathlib import Path
import ionworkspipeline as iwp
import pandas as pd
import iwutil
We first load in the data and the previously fitted MSMR parameters
# Load data
gitt_data = pd.read_csv(Path("synthetic_data") / "full_cell" / "gitt.csv")
# Extract relaxed voltages from GITT to get OCP data
ocp_data = iwp.data_fits.preprocess.pulse_data_to_ocp(gitt_data)
# Read in previously fitted parameters
anode_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "negative_electrode_ocp.json"
)
cathode_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "positive_electrode_ocp.json"
)
Next we choose the parameters to fit. Here we fit the lower and upper excess capacities in each electrode. We create iwp.Parameter objects for the parameters we want to fit, and set the initial guess and bounds for the parameters.
# Fit lower and upper excess capacities
# also add the electrode capacities to the parameters
Q_use = ocp_data["Capacity [A.h]"].max()
Q_n_lowex = iwp.Parameter(
"Q_n_lowex", initial_value=0.01 * Q_use, bounds=(0, 0.2 * Q_use)
)
Q_p_lowex = iwp.Parameter("Q_p_lowex", initial_value=0.1 * Q_use, bounds=(0, Q_use))
Q_n_uppex = iwp.Parameter(
"Q_n_uppex", initial_value=0.1 * Q_use, bounds=(0, 0.2 * Q_use)
)
Q_p_uppex = iwp.Parameter("Q_p_uppex", initial_value=0.5 * Q_use, bounds=(0, Q_use))
parameters = {
"Negative electrode lower excess capacity [A.h]": Q_n_lowex,
"Positive electrode lower excess capacity [A.h]": Q_p_lowex,
"Negative electrode upper excess capacity [A.h]": Q_n_uppex,
"Positive electrode upper excess capacity [A.h]": Q_p_uppex,
"Negative electrode capacity [A.h]": Q_n_lowex + Q_use + Q_n_uppex,
"Positive electrode capacity [A.h]": Q_p_lowex + Q_use + Q_p_uppex,
"Usable capacity [A.h]": Q_use,
}
Next we construct the objective function and data fit object
# Construct objective function and data fit for electrode balance
objective = iwp.objectives.MSMRFullCell(
ocp_data,
options={
# these are the voltage ranges over which to evaluate the half-cell OCP curves
"negative voltage limits": (0, 2),
"positive voltage limits": (2.5, 5),
},
)
electrode_balance = iwp.DataFit(
objective,
parameters=parameters,
)
# Construct a pipeline element to calculate the electrode stoichiometry windows for the
# full-cell based on the electrode total, lower and upper excess capacities
sto_windows = iwp.calculations.StoichiometryLimitsFromCapacity()
and put them into a pipeline
# Construct pipeline to load MSMR parameters and fit electrode balance
pipeline = iwp.Pipeline(
{
"temperature": iwp.direct_entries.temperatures(298.15),
"anode MSMR": anode_params,
"cathode MSMR": cathode_params,
"electrode balance": electrode_balance,
"stoichiometry windows": sto_windows,
}
)
params_fit = pipeline.run()
/home/docs/checkouts/readthedocs.org/user_builds/ionworks-ionworkspipeline/envs/v0.3.6/lib/python3.11/site-packages/pybamm/expression_tree/functions.py:152: RuntimeWarning: overflow encountered in exp
return self.function(*evaluated_children)
We save the results for use in the following notebook
# Save full cell balance to json
params_to_save_list = [
"Negative electrode lower excess capacity [A.h]",
"Negative electrode capacity [A.h]",
"Negative electrode upper excess capacity [A.h]",
"Negative electrode stoichiometry at minimum SOC",
"Negative electrode stoichiometry at maximum SOC",
"Positive electrode lower excess capacity [A.h]",
"Positive electrode capacity [A.h]",
"Positive electrode upper excess capacity [A.h]",
"Positive electrode stoichiometry at minimum SOC",
"Positive electrode stoichiometry at maximum SOC",
"Usable capacity [A.h]",
]
params_to_save = {k: v for k, v in params_fit.items() if k in params_to_save_list}
iwutil.save.json(params_to_save, Path("fitted_parameters") / "full_cell_balance.json")
Finally, we plot the results
electrode_balance.plot_fit_results()
electrode_balance.plot_trace()
/home/docs/checkouts/readthedocs.org/user_builds/ionworks-ionworkspipeline/envs/v0.3.6/lib/python3.11/site-packages/ionworkspipeline/data_fits/data_fit.py:586: UserWarning: The figure layout has changed to tight
fig.tight_layout()
(<Figure size 1200x700 with 6 Axes>,
array([<Axes: xlabel='Iteration', ylabel='Cost'>,
<Axes: xlabel='Iteration', ylabel='Q_n_lowex'>,
<Axes: xlabel='Iteration', ylabel='Q_n_uppex'>,
<Axes: xlabel='Iteration', ylabel='Q_p_lowex'>,
<Axes: xlabel='Iteration', ylabel='Q_p_uppex'>, <Axes: >],
dtype=object))