4. Full-cell validation¶
In this example we load in the previous results from OCP fitting, electrode balancing and half-cell GITT fitting to create a parameter set for the full cell. We then use validate this parameter set against the real parameters.
from pathlib import Path
import ionworkspipeline as iwp
import pybamm
from plots import constant_current, drive_cycle
from true_parameters.parameters import full_cell
We begin by making a list of “known parameters”. These are values that are already known from other sources (e.g. spec sheet, direct measurement, other experiments). For this example we load the known parameters from the true parameters used to generate the synthetic data.
# Known values
KNOWN_PARAMETERS = [
# Negative electrode
"Negative electrode thickness [m]",
"Negative electrode conductivity [S.m-1]",
"Negative electrode porosity",
"Negative electrode active material volume fraction",
"Negative particle radius [m]",
"Negative electrode OCP entropic change [V.K-1]",
# Separator
"Separator thickness [m]",
"Separator porosity",
# Positive electrode
"Positive electrode thickness [m]",
"Positive electrode conductivity [S.m-1]",
"Positive electrode porosity",
"Positive electrode active material volume fraction",
"Positive particle radius [m]",
"Positive electrode OCP entropic change [V.K-1]",
# Cell
"Electrode height [m]",
"Electrode width [m]",
# these are user-specified at the cell level and may differ from the actual
# voltage and capacity as measured in the experiment
"Lower voltage cut-off [V]",
"Upper voltage cut-off [V]",
"Open-circuit voltage at 0% SOC [V]",
"Open-circuit voltage at 100% SOC [V]",
"Nominal cell capacity [A.h]",
"Current function [A]",
]
Next we load in the known parameters
true_parameter_values = full_cell()
known_parameter_values = {k: true_parameter_values[k] for k in KNOWN_PARAMETERS}
known_values = iwp.direct_entries.DirectEntry(
known_parameter_values, "Measured or assumed values"
)
and load in the previously fitted open circuit, electrode balancing and GITT parameters
ocp_n = iwp.calculations.ocp_data_interpolant_from_csv(
"negative",
"fitted_parameters/negative_electrode_ocp.csv",
)
ocp_p = iwp.calculations.ocp_data_interpolant_from_csv(
"positive",
"fitted_parameters/positive_electrode_ocp.csv",
)
electrode_balance_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "full_cell_balance.json",
)
def j0_n(c_e, c_s_surf, c_s_max, T):
j0_ref = pybamm.Parameter(
"Negative electrode reference exchange-current density [A.m-2]"
)
alpha = 0.5
c_e_ref = 1000
return (
j0_ref
* (c_e / c_e_ref) ** (1 - alpha)
* (c_s_surf / c_s_max) ** alpha
* (1 - c_s_surf / c_s_max) ** (1 - alpha)
)
def j0_p(c_e, c_s_surf, c_s_max, T):
j0_ref = pybamm.Parameter(
"Positive electrode reference exchange-current density [A.m-2]"
)
alpha = 0.5
c_e_ref = 1000
return (
j0_ref
* (c_e / c_e_ref) ** (1 - alpha)
* (c_s_surf / c_s_max) ** alpha
* (1 - c_s_surf / c_s_max) ** (1 - alpha)
)
exchange_current_params = iwp.direct_entries.DirectEntry(
{
"Negative electrode exchange-current density [A.m-2]": j0_n,
"Positive electrode exchange-current density [A.m-2]": j0_p,
},
source="Known functional forms for exchange-current densities",
)
negative_gitt_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "negative_electrode_gitt.json",
)
positive_gitt_params = iwp.direct_entries.from_json(
Path("fitted_parameters") / "positive_electrode_gitt.json",
)
Next we construct a pipeline that uses the known parameters and the fitted parameters to create a full cell parameter set.
pipeline = iwp.Pipeline(
{
"known values": known_values,
"negative OCP": ocp_n,
"positive OCP": ocp_p,
"Electrode balance parameters": electrode_balance_params,
"Exchange-current densities": exchange_current_params,
"Negative GITT parameters": negative_gitt_params,
"Positive GITT parameters": positive_gitt_params,
"Defaults": iwp.direct_entries.standard_defaults(),
"Bruggeman": iwp.direct_entries.bruggeman(),
"Temperatures": iwp.direct_entries.temperatures(298.15),
"Electrolyte": iwp.direct_entries.nyman_electrolyte(1000),
"Maximum concentration negative": iwp.calculations.ElectrodeCapacity(
"negative",
unknown="maximum concentration",
method="capacity",
),
"Maximum concentration positive": iwp.calculations.ElectrodeCapacity(
"positive",
unknown="maximum concentration",
method="capacity",
),
"Initial concentration negative": iwp.calculations.InitialSOCHalfCell(
"negative", "maximum"
),
"Initial concentration positive": iwp.calculations.InitialSOCHalfCell(
"positive", "maximum"
),
}
)
We run the pipeline
fitted_parameter_values = pipeline.run()
and export a python script for the full cell parameter values
# Export a .py file with the PyBaMM parameters
# Note: if we specify data_path=None, the variable DATA_PATH will be set to the
# directory containing the exported script
iwp.util.export_python_script(
fitted_parameter_values,
"fitted_parameters/full_cell.py",
data_path=None,
)
Finally, we validate this parameter set against the known parameters.
model = pybamm.lithium_ion.SPMe()
true_parameter_values = pybamm.ParameterValues(true_parameter_values)
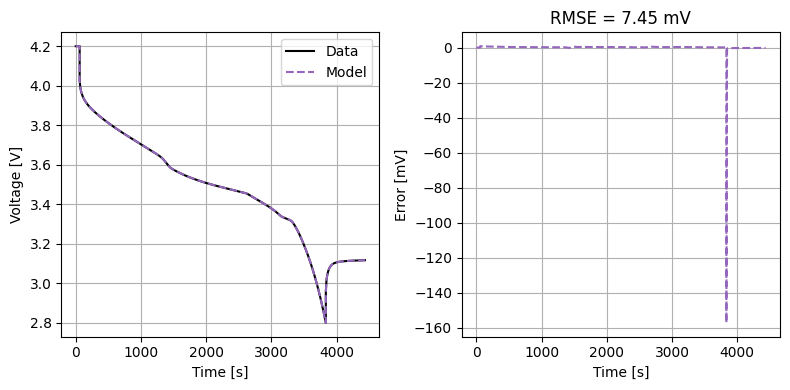
def plot_constant_current(model, Crate=1):
constant_current(
model,
true_parameter_values,
fitted_parameter_values,
Crate=Crate,
initial_soc=1,
)
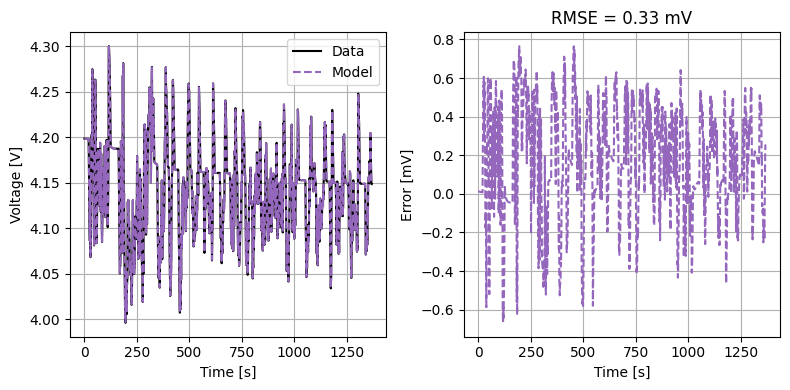
def plot_drive_cycle(model, peak_current=None):
peak_current = peak_current or true_parameter_values["Nominal cell capacity [A.h]"]
drive_cycle(
model,
true_parameter_values,
fitted_parameter_values,
peak_current=peak_current,
initial_soc=1,
)
plot_constant_current(model, Crate=1)
plot_drive_cycle(model)