Data Fit example¶
This notebook shows how the DataFit class works via examples. For design principles, see the user_guide
Simple non-battery example¶
The Ionworks battery parameter pipeline is built around PyBaMM models, so we start by creating a model which we will then use to generate and fit synthetic data
import pybamm
import numpy as np
import pandas as pd
import ionworkspipeline as iwp
class LotkaVolterra(pybamm.BaseModel):
def __init__(self):
super().__init__()
x = pybamm.Variable("x")
y = pybamm.Variable("y")
a = pybamm.Parameter("a")
b = pybamm.Parameter("b")
c = pybamm.Parameter("c")
d = pybamm.Parameter("d")
self.rhs = {x: a * x - b * x * y, y: -c * y + d * x * y}
self.initial_conditions = {x: 2, y: 1}
self.variables = {"x": x, "y": y}
model = LotkaVolterra()
true_inputs = {"a": 1.5, "b": 0.5, "c": 3.0, "d": 0.3}
true_params = pybamm.ParameterValues(true_inputs)
true_params.process_model(model)
solver = pybamm.ScipySolver()
t_data = np.linspace(0, 10, 1000)
solution = solver.solve(model, t_data)
# extract data and add noise
x_data = solution["x"].data + 0.1 * np.random.rand(t_data.size)
y_data = solution["y"].data + 0.1 * np.random.rand(t_data.size)
data = pd.DataFrame({"t": t_data, "x": x_data, "y": y_data})
To fit this model we set up a custom Objective class which defines how the error function is calculated from the solution
class Objective(iwp.objectives.Objective):
def process_data(self):
data = self.data["data"]
self._processed_data = {"x": data["x"], "y": data["y"]}
def build(self, all_parameter_values):
data = self.data["data"]
t_data = data["t"].values
model = LotkaVolterra()
all_parameter_values.process_model(model)
solver = pybamm.CasadiSolver()
def run(inputs, full_output=False):
sol = solver.solve(model, t_data, inputs=inputs)
return {"x": sol["x"].data, "y": sol["y"].data}
self.run_ = run
objective = Objective(data)
We can then specify which parameters need to be fitted, which parameters are known (in this case none), define bounds, and pass this to the standard DataFit class to fit the unknown parameters
fit_parameters = {
"a": iwp.Parameter("a", bounds=(0, np.inf)),
"b": iwp.Parameter("b", bounds=(0, np.inf)),
"c": iwp.Parameter("c", bounds=(0, np.inf)),
"d": iwp.Parameter("d", bounds=(0, np.inf)),
}
known_parameters = {}
optimizer = iwp.optimizers.ScipyLeastSquares(verbose=2)
data_fit = iwp.DataFit(objective, parameters=fit_parameters, optimizer=optimizer)
new_parameter_values = data_fit.run(known_parameters)
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 2.1200e+03 5.58e+02
1 3 1.8888e+03 2.31e+02 3.56e-01 4.45e+02
2 4 1.6359e+03 2.53e+02 7.06e-01 2.86e+03
3 5 1.2493e+03 3.87e+02 3.95e-01 1.70e+03
4 6 2.2818e+02 1.02e+03 7.10e-01 6.66e+03
5 8 9.7682e+01 1.30e+02 2.28e-01 1.72e+03
6 9 6.9182e+01 2.85e+01 4.62e-01 6.87e+03
7 10 1.5377e+00 6.76e+01 2.60e-01 9.08e+02
8 11 2.0937e-01 1.33e+00 1.75e-02 2.39e+01
9 12 2.0874e-01 6.28e-04 5.15e-04 8.31e-02
10 13 2.0874e-01 6.44e-09 3.37e-07 3.44e-04
11 14 2.0874e-01 2.01e-13 1.21e-08 3.53e-06
Both `ftol` and `xtol` termination conditions are satisfied.
Function evaluations 14, initial cost 2.1200e+03, final cost 2.0874e-01, first-order optimality 3.53e-06.
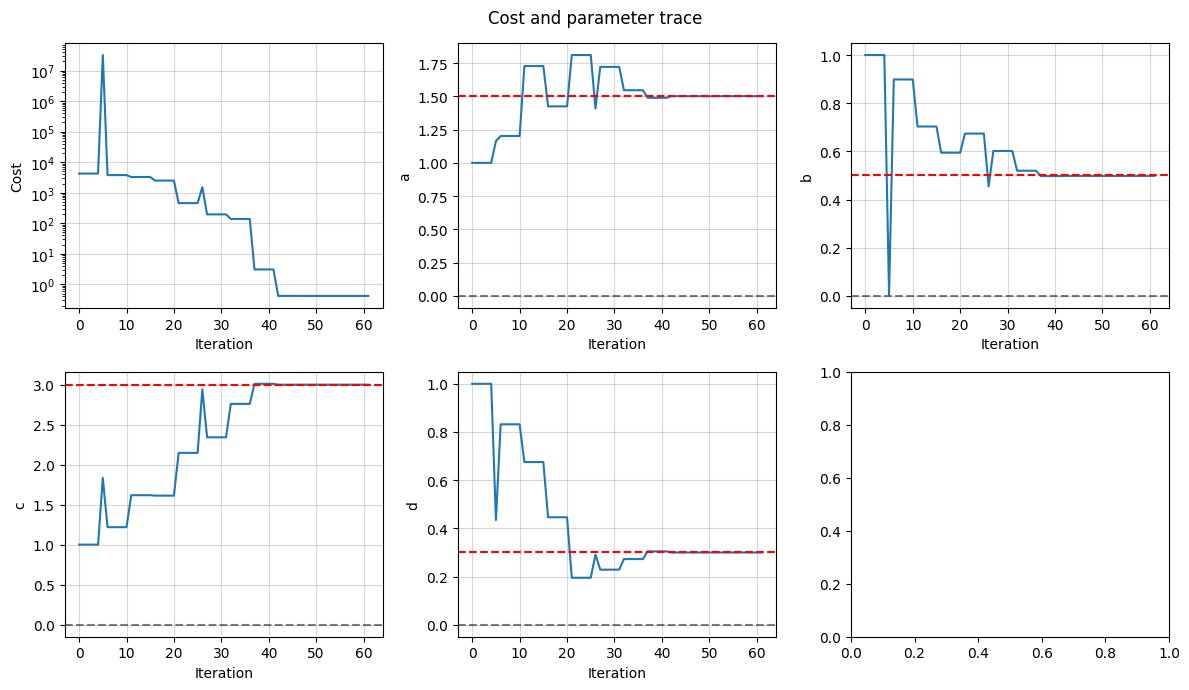
We can plot the trace of the cost and the parameter values to see how the fit progressed
fig, axes = data_fit.plot_trace()
for ax, true_value in zip(axes[1:], true_inputs.values()):
ax.axhline(true_value, color="r", linestyle="--")
/home/docs/checkouts/readthedocs.org/user_builds/ionworks-ionworkspipeline/envs/v0.3.6/lib/python3.11/site-packages/ionworkspipeline/data_fits/data_fit.py:586: UserWarning: The figure layout has changed to tight
fig.tight_layout()
We can reuse the same Objective and DataFit class but change which parameters are known and which are to be fitted. This flexibility is useful for battery models where we want to fit only a subset of the parameters to experimental data while keeping other parameters fixed.
fit_parameters = {
"a": iwp.Parameter("a", bounds=(0, np.inf)),
"b": iwp.Parameter("b", bounds=(0, np.inf)),
}
known_parameters = {"c": 3, "d": 0.3}
data_fit = iwp.DataFit(objective, parameters=fit_parameters, optimizer=optimizer)
new_parameter_values = data_fit.run(known_parameters)
for k, v in new_parameter_values.items():
true = true_inputs[k]
print(f"Parameter {k}: true: {true:.3f}, fitted: {v:.3f}")
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 2.6449e+03 9.63e+02
1 2 2.5971e+03 4.79e+01 5.15e-01 1.04e+03
2 3 2.5814e+03 1.57e+01 3.69e-02 1.26e+03
3 4 2.5577e+03 2.37e+01 1.94e-02 1.51e+03
4 5 2.5214e+03 3.64e+01 2.23e-02 1.82e+03
5 6 2.4645e+03 5.69e+01 2.99e-02 2.21e+03
6 7 2.3725e+03 9.20e+01 4.23e-02 2.73e+03
7 8 2.2127e+03 1.60e+02 6.29e-02 3.51e+03
8 9 1.9036e+03 3.09e+02 1.00e-01 4.76e+03
9 10 1.2573e+03 6.46e+02 1.77e-01 6.05e+03
10 11 3.5632e+02 9.01e+02 3.33e-01 9.43e+02
11 12 3.1341e+01 3.25e+02 3.66e-01 3.32e+03
12 13 2.7649e-01 3.11e+01 1.84e-02 7.93e+01
13 14 2.0989e-01 6.66e-02 2.68e-03 1.96e+00
14 15 2.0988e-01 1.01e-05 1.14e-05 7.08e-03
15 16 2.0988e-01 1.29e-10 3.89e-08 1.91e-05
`ftol` termination condition is satisfied.
Function evaluations 16, initial cost 2.6449e+03, final cost 2.0988e-01, first-order optimality 1.91e-05.
Parameter a: true: 1.500, fitted: 1.500
Parameter b: true: 0.500, fitted: 0.498
Battery example: constant-current experiment¶
We now repeat the same procedure for a battery model. We start by creating a model and solving it to generate synthetic data
model = pybamm.lithium_ion.SPM()
parameter_values = pybamm.ParameterValues("Chen2020")
sim = pybamm.Simulation(model, parameter_values=parameter_values)
sim.solve(np.linspace(0, 3600, 1000))
data = pd.DataFrame(
{x: sim.solution[x].entries for x in ["Time [s]", "Current [A]", "Voltage [V]"]}
)
Now we set up which parameters are to be fitted, and call the pre-defined CurrentDriven objective function, which solves the model with the current from the data and calculates the error between voltage from the model and the data
# In this example we just fit the diffusivity in the positive electrode
parameters = {
"Positive particle diffusivity [m2.s-1]": iwp.Parameter("D_s", initial_value=1e-15),
}
objective = iwp.objectives.CurrentDriven(data, options={"model": model})
current_driven = iwp.DataFit(objective, parameters=parameters)
# make sure we're not accidentally initializing with the correct values by passing
# them in
params_for_pipeline = {k: v for k, v in parameter_values.items() if k not in parameters}
params_fit = current_driven.run(params_for_pipeline)
print(
f"True parameter value: {parameter_values['Positive particle diffusivity [m2.s-1]']:.3e}"
)
print(
f"Fitted parameter value: {params_fit['Positive particle diffusivity [m2.s-1]']:.3e}"
)
[IDAS ERROR] IDACalcIC
The linesearch algorithm failed: step too small or too many backtracks.
[IDAS ERROR] IDASolve
At t = 0 and h = 8.88112e-60, the corrector convergence failed repeatedly or with |h| = hmin.
True parameter value: 4.000e-15
Fitted parameter value: 4.000e-15
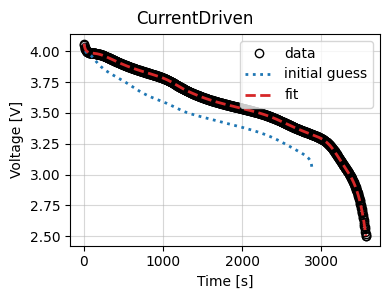
We can also plot the fit results
current_driven.plot_fit_results()
{'CurrentDriven': [(<Figure size 400x300 with 1 Axes>,
<Axes: xlabel='Time [s]', ylabel='Voltage [V]'>)]}