Exchange-current density from resistance¶
In this example we show how to fit the exchange-current density to resistance data calculated from synthetic pulse data.
import pybamm
import ionworkspipeline as iwp
import pandas as pd
We begin by generating synthetic pulse data for a half-cell. In this example we use the standard symmetric exchange-current density function and will fit the reference value. We set up the model and parameters
model = pybamm.lithium_ion.SPMe(
{"working electrode": "positive", "particle": "uniform profile"}
)
model.events = []
parameter_values = pybamm.ParameterValues("Xu2019")
def j0(c_e, c_s_surf, c_s_max, T):
j0_ref = pybamm.Parameter(
"Positive electrode reference exchange-current density [A.m-2]"
)
c_e_init = pybamm.Parameter("Initial concentration in electrolyte [mol.m-3]")
return (
j0_ref
* (c_e / c_e_init) ** 0.5
* (c_s_surf / c_s_max) ** 0.5
* (1 - c_s_surf / c_s_max) ** 0.5
)
parameter_values.update(
{
"Positive electrode reference exchange-current density [A.m-2]": 5,
"Positive electrode exchange-current density [A.m-2]": j0,
},
check_already_exists=False,
)
and then simulate a GITT experiment
C_rate = 1 / 15
pause_s = 10 # 10s
step_s = 5 * 60 # 5m
step_h = step_s / 3600
rest_s = 1 * 3600 # 1h
max_cycles = int(1 / C_rate / step_h * 2)
V_min = parameter_values["Lower voltage cut-off [V]"]
gitt_experiment = pybamm.Experiment(
[
(
pybamm.step.rest(pause_s, period=1),
pybamm.step.c_rate(
C_rate,
duration=step_s,
period=step_s / 100,
termination=f"{V_min} V",
),
pybamm.step.rest(rest_s, period=rest_s / 100),
),
]
)
sim = pybamm.Simulation(
model, parameter_values=parameter_values, experiment=gitt_experiment
)
sol = None
i = 0
# Run until the voltage drops below the lower cut-off
# during step 1 (the discharge step)
while sol is None or sol.cycles[-1].steps[1].termination == "final time":
sol = sim.solve(starting_solution=sol)
i += 1
if i > max_cycles:
raise ValueError("Reached maximum number of cycles")
We export the solution data to a dataframe. Note: for synthetic data we can directly store the stoichiometry, whereas in practice we would need to compute this from the half-cell capacity and the charge passed.
variables = [
"Time [s]",
"Current [A]",
"Voltage [V]",
"Positive electrode stoichiometry",
]
df = pd.DataFrame(sim.solution.get_data_dict(variables))
df = df.rename(columns={"Cycle": "Cycle number", "Step": "Step number"})
Next we calculate the 1s resistance from the solution data using the function iwp.objectives.pulse.calculate_pulse_resistance. We pass in the step number “1” which is the “on” step of our pulse data (steps 0 and 2 are the “off” steps).
steps = [1]
dts = [1]
data = iwp.objectives.pulse.calculate_pulse_resistance("positive", df, steps, dts)
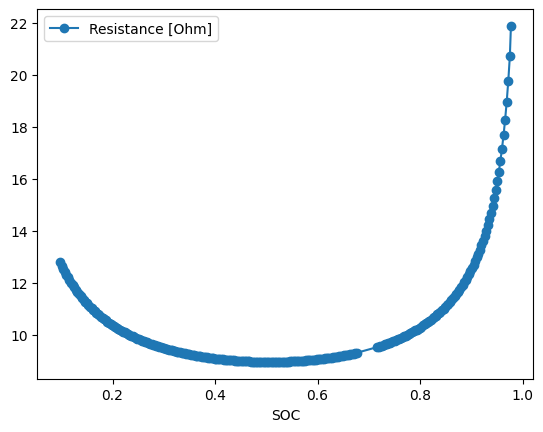
We can plot the calculated resistance as a function of stoichiometry
data.plot(x="SOC", y="Resistance [Ohm]", style="o-")
<Axes: xlabel='SOC'>
Next we set up the objective that will be used to perform the fit
model = iwp.objectives.HalfCellResistanceModel("positive")
objective = iwp.objectives.Resistance(data, {"model": model})
we fit the exchange-current density and an extra scalar “contact resistance” parameter that accounts for any other resistances
parameters = {
"Positive electrode reference exchange-current density [A.m-2]": iwp.Parameter(
"j0_ref", initial_value=1, bounds=(0.1, 10)
),
"Contact resistance [Ohm]": iwp.Parameter("R_contact"),
}
data_fit = iwp.DataFit(objective, parameters=parameters)
We pass in the known parameter values and run the fit
# make sure we're not accidentally initializing with the correct values by passing
# them in
params_for_pipeline = {k: v for k, v in parameter_values.items() if k not in parameters}
data_fit.run(params_for_pipeline)
{'Positive electrode reference exchange-current density [A.m-2]': 5.047136271614368,
'Contact resistance [Ohm]': 2.6525385581252463}
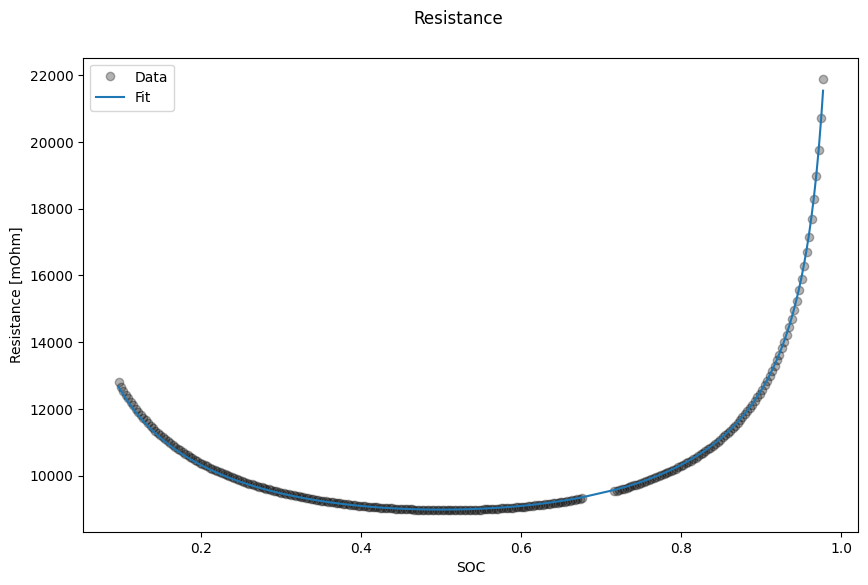
Finally we plot the results
data_fit.plot_fit_results()
{'Resistance': [(<Figure size 1000x600 with 1 Axes>,
<Axes: xlabel='SOC', ylabel='Resistance [mOhm]'>)]}