Particle Swarm Optimizer¶
In this example, we will use the Particle Swarm Optimizer (PSO) to fit a battery model to some synthetic data.
We begin by generating some synthetic data from a battery model.
import pybamm
import ionworkspipeline as iwp
import numpy as np
import pandas as pd
# Load the model and parameter values
model = pybamm.lithium_ion.SPM()
parameter_values = pybamm.ParameterValues("Chen2020")
parameter_values.update(
{
"Negative electrode active material volume fraction": 0.75,
"Positive electrode active material volume fraction": 0.665,
}
)
# Generate the data
t_eval = np.arange(0, 900, 3)
sim = pybamm.Simulation(model, parameter_values=parameter_values)
sol = sim.solve(t_eval)
data = pd.DataFrame(
{x: sim.solution[x].entries for x in ["Time [s]", "Current [A]", "Voltage [V]"]}
)
# add noise to the data
sigma = 0.001
data["Voltage [V]"] += np.random.normal(0, sigma, len(data))
Next we define the parameters to fit, along with initial guesses and bounds.
parameters = {
"Negative electrode active material volume fraction": iwp.Parameter(
"Negative electrode active material volume fraction",
initial_value=0.5,
bounds=(0, 1),
),
"Positive electrode active material volume fraction": iwp.Parameter(
"Positive electrode active material volume fraction",
initial_value=0.5,
bounds=(0, 1),
),
}
We set up the objective function, which in this case is the current driven objective. This takes the time vs current data and runs the model, comparing the model’s predictions for the voltage with the experimental data.
objective = iwp.objectives.CurrentDriven(data, options={"model": model})
Then we set up the DataFit object, which takes the objective function, the parameters to fit, and the optimizer. Here we use the Particle Swarm Optimizer (PSO) from the pints library and set the maximum number of iterations to 100.
optimizer = iwp.optimizers.Pints(max_iterations=100, method="PSO")
data_fit = iwp.DataFit(objective, parameters=parameters, optimizer=optimizer)
Next we run the data fit, passing the parameters that are not being fit as a dictionary.
params_for_pipeline = {k: v for k, v in parameter_values.items() if k not in parameters}
params_fit = data_fit.run(params_for_pipeline)
for k, v in params_fit.items():
print(f"{k}: {parameter_values[k]:.3e} (true) {v:.3e} (fit)")
Negative electrode active material volume fraction: 7.500e-01 (true) 7.419e-01 (fit)
Positive electrode active material volume fraction: 6.650e-01 (true) 6.706e-01 (fit)
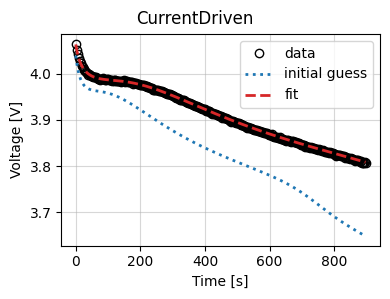
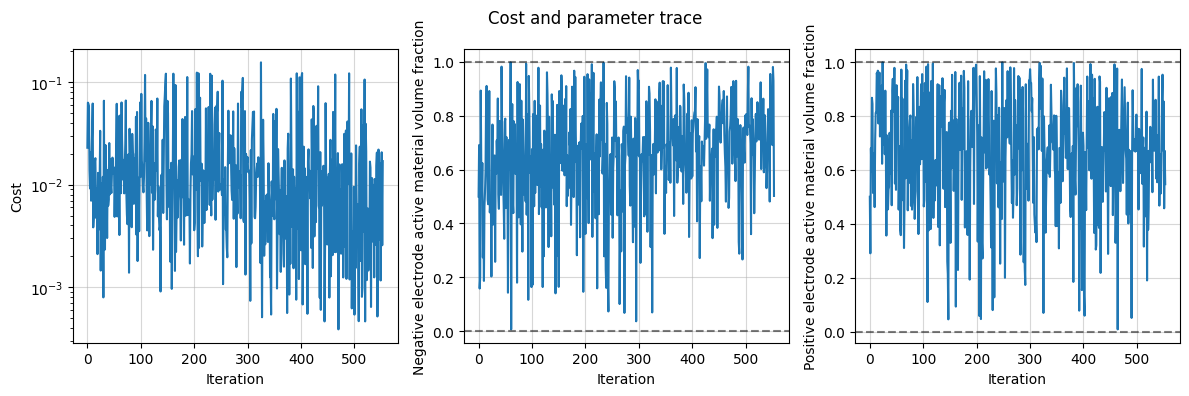
Finally, we plot the results and the trace of the cost function and parameter values.
data_fit.plot_fit_results()
data_fit.plot_trace()
(<Figure size 1200x400 with 3 Axes>,
array([<Axes: xlabel='Iteration', ylabel='Cost'>,
<Axes: xlabel='Iteration', ylabel='Negative electrode active material volume fraction'>,
<Axes: xlabel='Iteration', ylabel='Positive electrode active material volume fraction'>],
dtype=object))