Fitting OCP hysteresis¶
In this example, we show how to fit the lithiation and delithiation OCPs at the same time in a way that is consistent with the MSMR model. In particular, we want the total capacity to be the same during lithiation and delithiation. To ensure this, we fit a single capacity for each species, and separate reference potential and activities for lithiation and delithiation (5n parameters for n species).
import ionworkspipeline as iwp
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.signal import savgol_filter
We use data from half-cells constructed from an LGM50 cell, as reported in https://github.com/paramm-team/pybamm-param
data = {}
Q_max_data = 0
fig, ax = plt.subplots(1, 3, figsize=(10, 5))
for direction in ["lithiation", "delithiation"]:
# Smooth
raw_ocp_data = pd.read_csv(f"anode_ocp_{direction}.csv", comment="#")
q_smoothed = savgol_filter(
raw_ocp_data["Capacity [A.h]"].values, window_length=11, polyorder=1
)
U_smoothed = savgol_filter(
raw_ocp_data["Voltage [V]"].values, window_length=11, polyorder=1
)
# Store in a dataframe
ocp_data = pd.DataFrame({"Capacity [A.h]": q_smoothed, "Voltage [V]": U_smoothed})
# Preprocess
ocp_data = iwp.data_fits.preprocess.sort_capacity_and_ocp(ocp_data)
ocp_data = iwp.data_fits.preprocess.remove_ocp_extremes(ocp_data)
q = ocp_data["Capacity [A.h]"].values
U = ocp_data["Voltage [V]"].values
data[direction] = ocp_data
# Maximum capacity, will be used later as an initial guess for the electrode capacity
Q_max_data = max(Q_max_data, ocp_data["Capacity [A.h]"].max())
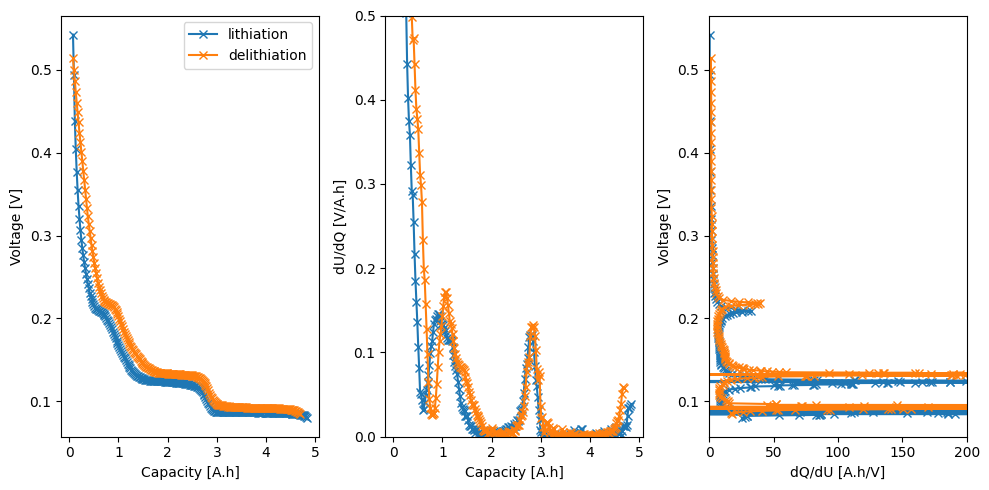
# Plot
dUdQ = -np.gradient(U, q)
dQdU = -np.gradient(q, U)
ax[0].plot(q, U, "-x", label=direction)
ax[0].set_xlabel("Capacity [A.h]")
ax[0].set_ylabel("Voltage [V]")
ax[1].plot(q, dUdQ, "-x")
ax[1].set_xlabel("Capacity [A.h]")
ax[1].set_ylabel("dU/dQ [V/A.h]")
ax[1].set_ylim(0, 0.5)
ax[2].plot(dQdU, U, "-x")
ax[2].set_xlabel("dQ/dU [A.h/V]")
ax[2].set_ylabel("Voltage [V]")
ax[2].set_xlim(0, 200)
ax[0].legend()
fig.tight_layout()
Fit the lithiation and delithiation separately to get an initial guess and bounds for the parameters¶
See ocp_fitting.ipynb for details.
# Initial guess of Graphite from Verbrugge et al 2017 J. Electrochem. Soc. 164 E3243
msmr_param_init = iwp.Material("GRAPHITE_VERBRUGGE2017").parameter_values
msmr_param_fit = {}
for direction, data_ in data.items():
Q_use = data_["Capacity [A.h]"].max()
# Add Parameter objects for the MSMR parameters X_j, U0_j, and w_j
def bounds_function(var, initial_value, Q_total):
if var.startswith("Q"):
return 0, Q_total
elif var.startswith("X"):
return 0, 1
elif var.startswith("U"):
return max(0, initial_value - 0.1), initial_value + 0.1
elif var.startswith("w"):
return 1e-2, 100
msmr_params = {
**iwp.objectives.get_msmr_capacity_params_for_fit(
{"Q_n_lowex": 0.01 * Q_use},
"negative",
Q_use,
parameter_format="lower excess capacity only",
),
**iwp.objectives.get_msmr_params_for_fit(
msmr_param_init, "negative", Q_use, bounds_function=bounds_function
),
}
# Set up objective and fit
objective = iwp.objectives.MSMRHalfCell(
"negative", data_, options={"constrain_Xj": "false", "voltage limits": (0, 1.5)}
)
ocp_msmr = iwp.DataFit(objective, parameters=msmr_params)
# Run the fit
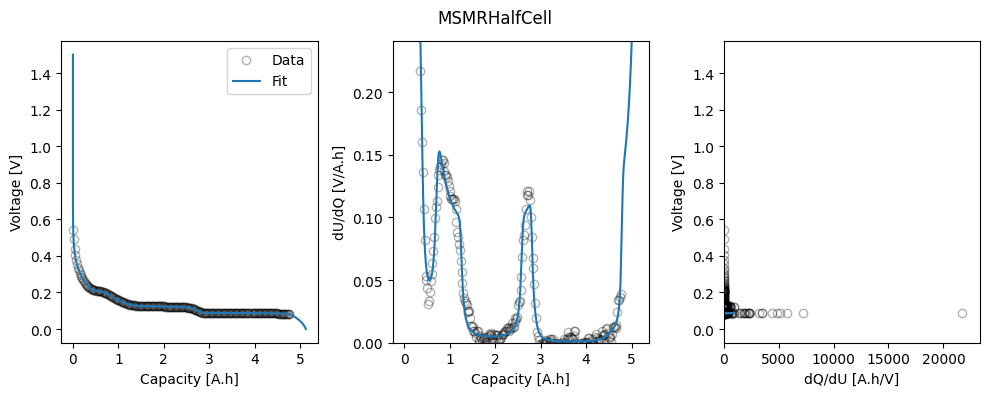
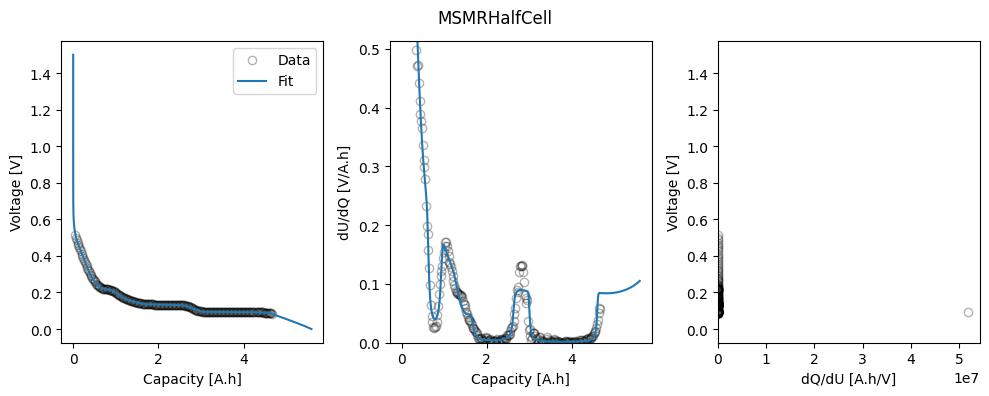
msmr_param_fit[direction] = ocp_msmr.run({"Ambient temperature [K]": 298.15})
fig_ax = ocp_msmr.plot_fit_results()["MSMRHalfCell"]
ax = fig_ax[0][1]
# ax[2].set_xlim(0, 200)
/home/docs/checkouts/readthedocs.org/user_builds/ionworks-ionworkspipeline/envs/v0.3.6/lib/python3.11/site-packages/pybamm/expression_tree/functions.py:152: RuntimeWarning: overflow encountered in exp
return self.function(*evaluated_children)
# Sort each one by Q so that we get capacities that are close to each other
# for the same electrode at the same time
msmr_param_fit_sorted = {
direction: iwp.objectives.msmr_sort_params(params, sort_by="Q_n")
for direction, params in msmr_param_fit.items()
}
# plot msmr param fit as a scatter plot on a log scale with the names as the x axis
# make sure each point has the same color in each direction
fig, ax = plt.subplots()
markers = {"delithiation": "o", "lithiation": "x"}
for direction, msmr_param in msmr_param_fit_sorted.items():
ax.set_prop_cycle(None)
msmr_param_sorted = dict(sorted(msmr_param.items()))
for name, value in msmr_param_sorted.items():
if "_0" in name:
ax.set_prop_cycle(None)
if "X_" in name:
continue
ax.scatter(name, value, label=direction, marker=markers[direction])
ax.set_yscale("log")
# rotate the x axis labels
_ = ax.set_xticklabels(ax.get_xticklabels(), rotation=45, ha="right")
/tmp/ipykernel_933/1576170936.py:16: UserWarning: set_ticklabels() should only be used with a fixed number of ticks, i.e. after set_ticks() or using a FixedLocator.
_ = ax.set_xticklabels(ax.get_xticklabels(), rotation=45, ha="right")
Fit the MSMR half-cell model with hysteresis¶
First we specify a custom callback that we will use to show the fit results on a single plot
class Callback(iwp.callbacks.SaveResultsCallback):
def __init__(self, folder=None):
self.folder = folder
self.data = []
self.fit_results = []
def on_objective_build(self, logs):
self.data.append(logs["data"])
def on_datafit_finish(self, logs):
self.fit_results.append(logs)
def plot_fit_results(self):
import matplotlib.pyplot as plt
colors = [{"data": "C0", "fit": "C0"}, {"data": "C1", "fit": "C1"}]
labels = [
{"data": "Data (delithiation)", "fit": "Fit (delithiation)"},
{"data": "Data (lithiation)", "fit": "Fit (lithiation)"},
]
fig, axes = plt.subplots(1, 3, figsize=(10, 4))
for idx, (data, fit_result) in enumerate(zip(self.data, self.fit_results)):
iwp.objectives.plot_half_cell_ocp(
data,
fit_result["outputs"],
fig_axes=(fig, axes),
colors=colors[idx],
labels=labels[idx],
)
fig.tight_layout()
return fig, axes
Then create objectives, parameters, and datafit, and run
def hysteresis_bounds_function(var, initial_value, Q_use):
if var.startswith("Q_") and "lowex" not in var:
return 0, Q_use
if "lowex" in var:
var = "Negative electrode lower excess capacity [A.h]"
var_lit = msmr_param_fit_sorted["lithiation"][var]
var_delit = msmr_param_fit_sorted["delithiation"][var]
min_var = min(var_lit, var_delit)
max_var = max(var_lit, var_delit)
delta_var = max_var - min_var
return min_var - 0.5 * delta_var, max_var + 0.5 * delta_var
average_param_init = {}
for var in msmr_param_fit_sorted["lithiation"]:
if "X_" in var:
continue
average_param_init[var] = np.mean(
[msmr_param_fit_sorted[direction][var] for direction in data]
)
objectives = {}
callback = Callback()
n_species = len([k for k in msmr_param_init if "U0_" in k])
for direction in ["lithiation", "delithiation"]:
q_fn = q_fn_delith = iwp.objectives.get_q_half_cell_msmr(
n_species, "negative", species_format="Qj", direction=direction
)
objectives[direction] = iwp.objectives.MSMRHalfCell(
"negative",
data[direction],
options={
"capacity function": q_fn,
"constrain_Xj": "false",
"dUdQ cutoff": 0.6,
"voltage limits": (0, 1.5),
},
callbacks=callback,
)
params_to_fit = {
**iwp.objectives.get_msmr_params_for_fit(
average_param_init,
"negative",
Q_max_data,
bounds_function=hysteresis_bounds_function,
species_format="Qj",
hysteresis=True,
),
}
# we fit the lower excess capacity for each direction
for direction in ["delithiation", "lithiation"]:
params_to_fit.update(
iwp.objectives.get_msmr_capacity_params_for_fit(
{
"Q_n_d_lowex": average_param_init[
"Negative electrode lower excess capacity [A.h]"
],
"Q_n_l_lowex": average_param_init[
"Negative electrode lower excess capacity [A.h]"
],
},
"negative",
Q_max_data,
parameter_format="lower excess capacity only",
direction=direction,
bounds_function=hysteresis_bounds_function,
)
)
data_fit = iwp.DataFit(objectives, parameters=params_to_fit)
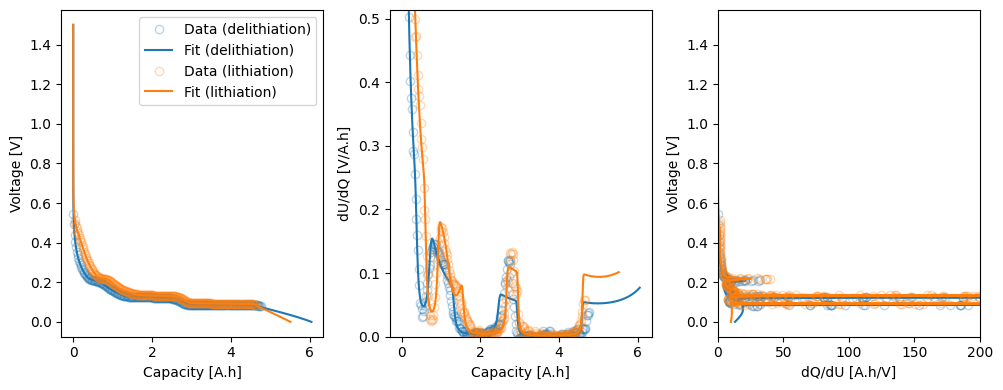
params_fit = data_fit.run({"Ambient temperature [K]": 298.15})
fig, axes = callback.plot_fit_results()
_ = axes[2].set_xlim(0, 200)
/home/docs/checkouts/readthedocs.org/user_builds/ionworks-ionworkspipeline/envs/v0.3.6/lib/python3.11/site-packages/pybamm/expression_tree/functions.py:152: RuntimeWarning: overflow encountered in exp
return self.function(*evaluated_children)